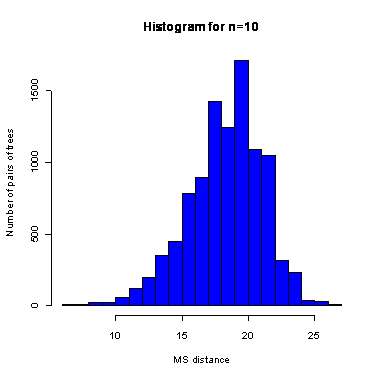
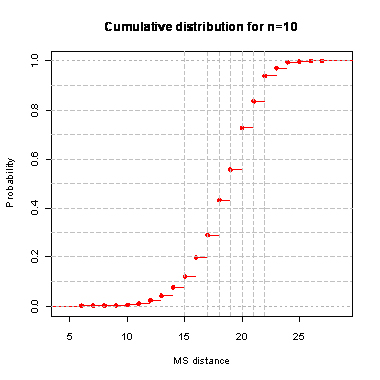
n=10
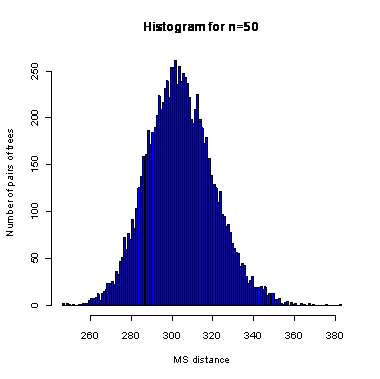
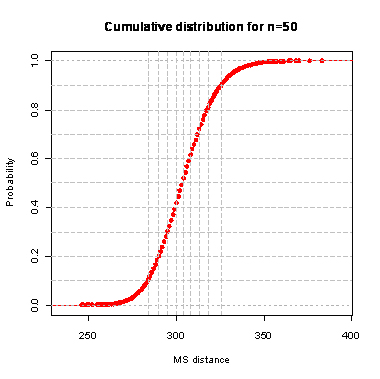
n=50
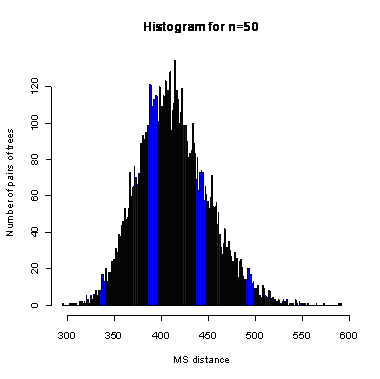
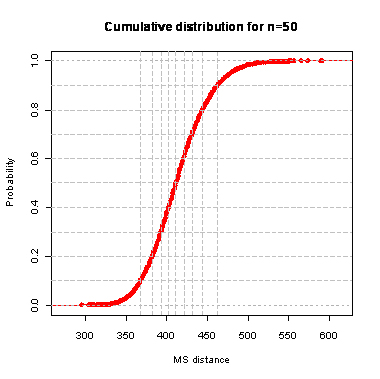
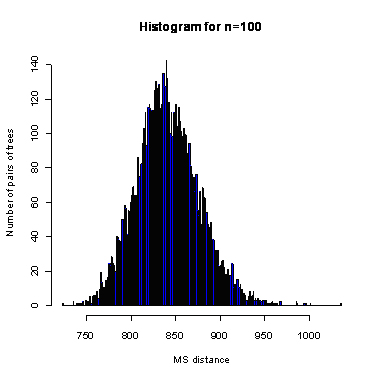
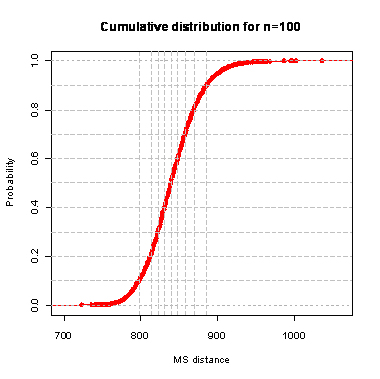
During the computation we analyzed 10000 pairs of trees that were chosen at random from among all possible unrooted binary trees with a particular number of leaves (n) under two the most popular models: the uniform model and the Yule model. In the case of the uniform model, an edge is added uniformly and randomly to any edge of the base tree. In this model, all phylogenetic trees have the same probability to be drawn. In the case of the Yule model, an edge is added uniformly and randomly to a pendant edge only, and the corresponding label is uniformly and randomly drawn as well.




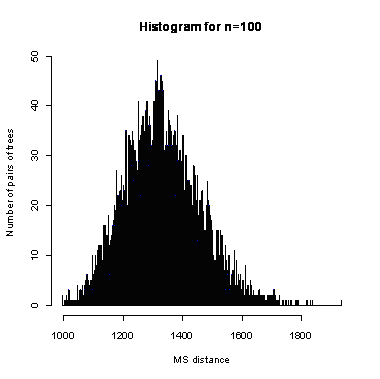
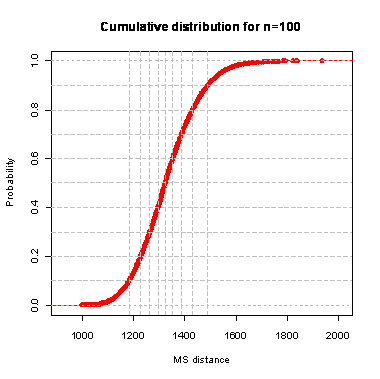
| n | Quantile | avg | sd | ||||||||||||
| 0.02 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 0.95 | 0.97 | |||
| 5 | 0 | 0 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 2.665 | 0.946 |
| 10 | 12 | 14 | 15 | 17 | 18 | 18 | 19 | 20 | 20 | 21 | 22 | 23 | 23 | 18.780 | 2.767 |
| 15 | 34 | 36 | 38 | 40 | 42 | 43 | 45 | 46 | 47 | 49 | 51 | 52 | 53 | 44.443 | 5.012 |
| 20 | 63 | 66 | 69 | 72 | 75 | 77 | 79 | 81 | 83 | 86 | 89 | 92 | 94 | 78.798 | 7.902 |
| 25 | 97 | 102 | 106 | 110 | 114 | 117 | 120 | 122 | 126 | 129 | 134 | 139 | 142 | 119.902 | 11.254 |
| 30 | 138 | 144 | 149 | 155 | 159 | 163 | 167 | 171 | 176 | 181 | 188 | 194 | 198 | 167.910 | 15.460 |
| 35 | 183 | 190 | 196 | 204 | 210 | 215 | 220 | 226 | 231 | 238 | 248 | 255 | 261 | 221.227 | 20.030 |
| 40 | 233 | 242 | 249 | 259 | 266 | 273 | 279 | 286 | 293 | 301 | 313 | 324 | 332 | 280.585 | 25.259 |
| 45 | 287 | 297 | 306 | 318 | 327 | 335 | 343 | 351 | 359 | 369 | 384 | 398 | 407 | 344.408 | 30.554 |
| 50 | 345 | 357 | 368 | 382 | 393 | 402 | 411 | 421 | 431 | 444 | 462 | 479 | 489 | 413.662 | 36.970 |
| 55 | 406 | 420 | 433 | 450 | 463 | 474 | 485 | 496 | 509 | 523 | 543 | 560 | 573 | 487.047 | 43.103 |
| 60 | 470 | 487 | 503 | 522 | 537 | 550 | 563 | 576 | 589 | 607 | 631 | 652 | 664 | 565.196 | 50.315 |
| 65 | 542 | 559 | 577 | 598 | 615 | 630 | 644 | 659 | 676 | 695 | 724 | 749 | 764 | 647.839 | 57.558 |
| 70 | 609 | 631 | 651 | 676 | 696 | 713 | 730 | 747 | 765 | 787 | 819 | 848 | 865 | 733.168 | 65.801 |
| 75 | 685 | 709 | 733 | 761 | 782 | 800 | 818 | 837 | 859 | 884 | 919 | 949 | 969 | 822.935 | 72.881 |
| 80 | 767 | 794 | 816 | 847 | 872 | 892 | 911 | 932 | 957 | 986 | 1029 | 1063 | 1088 | 918.048 | 82.411 |
| 85 | 847 | 878 | 903 | 937 | 964 | 987 | 1010 | 1034 | 1060 | 1093 | 1138 | 1177 | 1203 | 1016.300 | 92.055 |
| 90 | 933 | 964 | 994 | 1032 | 1061 | 1087 | 1112 | 1138 | 1165 | 1199 | 1249 | 1289 | 1317 | 1117.540 | 99.619 |
| 95 | 1021 | 1053 | 1085 | 1126 | 1157 | 1187 | 1214 | 1243 | 1277 | 1315 | 1369 | 1415 | 1445 | 1221.913 | 110.882 |
| 100 | 1108 | 1144 | 1181 | 1226 | 1263 | 1294 | 1322 | 1352 | 1386 | 1429 | 1487 | 1537 | 1572 | 1329.663 | 119.696 |
| 105 | 1201 | 1242 | 1281 | 1330 | 1369 | 1401 | 1434 | 1467 | 1503 | 1549 | 1612 | 1665 | 1705 | 1441.605 | 129.920 |
| 110 | 1300 | 1347 | 1385 | 1437 | 1478 | 1512 | 1547 | 1585 | 1623 | 1673 | 1742 | 1797 | 1838 | 1557.035 | 139.339 |
| 115 | 1394 | 1442 | 1487 | 1545 | 1589 | 1627 | 1664 | 1701 | 1747 | 1802 | 1877 | 1939 | 1981 | 1674.368 | 151.655 |
| 120 | 1494 | 1546 | 1592 | 1654 | 1702 | 1743 | 1784 | 1824 | 1871 | 1928 | 2013 | 2086 | 2133 | 1794.614 | 163.616 |
| 125 | 1597 | 1653 | 1705 | 1770 | 1820 | 1864 | 1911 | 1954 | 2005 | 2062 | 2152 | 2225 | 2275 | 1920.300 | 174.237 |
| 130 | 1705 | 1765 | 1820 | 1891 | 1945 | 1992 | 2039 | 2087 | 2140 | 2203 | 2293 | 2373 | 2425 | 2050.487 | 185.638 |
| 135 | 1811 | 1868 | 1933 | 2010 | 2067 | 2118 | 2164 | 2212 | 2268 | 2334 | 2431 | 2521 | 2578 | 2175.958 | 196.473 |
| 140 | 1924 | 1991 | 2050 | 2127 | 2188 | 2242 | 2298 | 2351 | 2409 | 2480 | 2587 | 2681 | 2740 | 2309.193 | 210.883 |
| 145 | 2043 | 2108 | 2170 | 2249 | 2316 | 2374 | 2430 | 2489 | 2552 | 2630 | 2734 | 2831 | 2890 | 2444.072 | 221.186 |
| 150 | 2153 | 2225 | 2295 | 2381 | 2452 | 2511 | 2570 | 2630 | 2696 | 2776 | 2894 | 2994 | 3060 | 2584.101 | 234.292 |
| 155 | 2265 | 2345 | 2420 | 2514 | 2585 | 2649 | 2707 | 2770 | 2841 | 2923 | 3051 | 3158 | 3225 | 2723.293 | 246.155 |
| 160 | 2385 | 2464 | 2541 | 2643 | 2724 | 2790 | 2856 | 2923 | 2996 | 3088 | 3213 | 3330 | 3405 | 2870.498 | 263.739 |
| 165 | 2505 | 2589 | 2673 | 2778 | 2862 | 2931 | 2998 | 3070 | 3151 | 3244 | 3387 | 3498 | 3574 | 3016.353 | 277.285 |
| 170 | 2641 | 2726 | 2807 | 2918 | 2998 | 3072 | 3145 | 3216 | 3297 | 3399 | 3545 | 3673 | 3758 | 3163.300 | 288.636 |
| 175 | 2748 | 2852 | 2944 | 3054 | 3143 | 3223 | 3298 | 3377 | 3461 | 3566 | 3721 | 3850 | 3936 | 3317.020 | 305.045 |
| 180 | 2894 | 2996 | 3091 | 3206 | 3292 | 3372 | 3448 | 3529 | 3618 | 3727 | 3894 | 4022 | 4111 | 3472.936 | 313.904 |
| 185 | 3007 | 3117 | 3214 | 3337 | 3434 | 3518 | 3602 | 3686 | 3783 | 3900 | 4066 | 4210 | 4298 | 3623.939 | 333.090 |
| 190 | 3154 | 3261 | 3360 | 3490 | 3590 | 3676 | 3760 | 3855 | 3957 | 4070 | 4245 | 4404 | 4512 | 3788.776 | 348.902 |
| 195 | 3276 | 3395 | 3499 | 3640 | 3747 | 3840 | 3929 | 4018 | 4124 | 4250 | 4430 | 4588 | 4694 | 3951.299 | 362.682 |
| 200 | 3427 | 3534 | 3648 | 3784 | 3889 | 3991 | 4090 | 4188 | 4301 | 4429 | 4614 | 4775 | 4879 | 4114.372 | 381.242 |
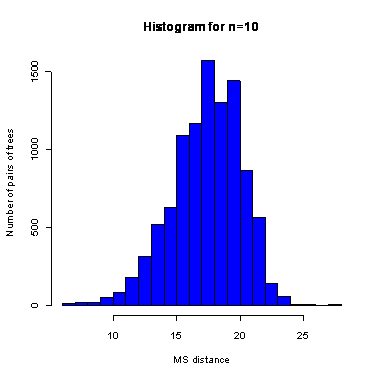
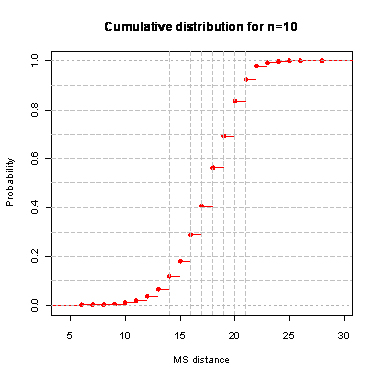
| n | Quantile | avg | sd | ||||||||||||
| 0.02 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 0.95 | 0.97 | |||
| 5 | 0 | 0 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 2.663 | 0.952 |
| 10 | 12 | 13 | 14 | 16 | 17 | 17 | 18 | 19 | 20 | 20 | 21 | 22 | 22 | 17.885 | 2.725 |
| 15 | 31 | 33 | 35 | 37 | 38 | 39 | 40 | 42 | 43 | 44 | 46 | 47 | 48 | 40.298 | 4.245 |
| 20 | 56 | 59 | 61 | 64 | 65 | 67 | 68 | 70 | 71 | 73 | 76 | 78 | 79 | 68.330 | 5.911 |
| 25 | 84 | 88 | 91 | 94 | 97 | 99 | 100 | 102 | 104 | 107 | 110 | 113 | 115 | 100.389 | 7.614 |
| 30 | 117 | 121 | 124 | 128 | 131 | 133 | 136 | 138 | 141 | 144 | 148 | 152 | 154 | 135.985 | 9.412 |
| 35 | 152 | 156 | 160 | 165 | 168 | 171 | 174 | 177 | 180 | 183 | 188 | 193 | 196 | 174.281 | 11.061 |
| 40 | 190 | 195 | 199 | 205 | 209 | 212 | 215 | 218 | 221 | 226 | 232 | 237 | 241 | 215.389 | 12.733 |
| 45 | 230 | 235 | 241 | 246 | 251 | 254 | 258 | 262 | 266 | 271 | 277 | 283 | 286 | 258.561 | 14.467 |
| 50 | 273 | 279 | 284 | 290 | 295 | 300 | 304 | 308 | 313 | 318 | 326 | 332 | 337 | 304.401 | 16.433 |
| 55 | 316 | 323 | 329 | 336 | 342 | 346 | 351 | 355 | 360 | 366 | 375 | 382 | 387 | 351.449 | 18.059 |
| 60 | 362 | 370 | 376 | 384 | 390 | 395 | 400 | 405 | 410 | 417 | 427 | 435 | 441 | 400.712 | 19.904 |
| 65 | 409 | 417 | 424 | 432 | 439 | 444 | 450 | 455 | 461 | 469 | 479 | 488 | 494 | 450.578 | 21.772 |
| 70 | 458 | 467 | 474 | 483 | 490 | 496 | 502 | 508 | 514 | 522 | 534 | 544 | 550 | 502.957 | 23.414 |
| 75 | 507 | 517 | 525 | 535 | 542 | 549 | 555 | 562 | 569 | 578 | 590 | 601 | 609 | 556.401 | 25.853 |
| 80 | 560 | 569 | 577 | 588 | 595 | 602 | 609 | 616 | 624 | 633 | 646 | 658 | 665 | 610.732 | 27.134 |
| 85 | 612 | 622 | 631 | 642 | 651 | 658 | 665 | 673 | 681 | 690 | 704 | 717 | 726 | 666.877 | 28.943 |
| 90 | 666 | 677 | 686 | 698 | 707 | 716 | 723 | 730 | 739 | 749 | 764 | 777 | 785 | 724.167 | 30.677 |
| 95 | 720 | 731 | 742 | 755 | 764 | 773 | 781 | 790 | 798 | 809 | 825 | 840 | 849 | 782.455 | 32.832 |
| 100 | 776 | 787 | 798 | 813 | 823 | 831 | 840 | 848 | 858 | 869 | 885 | 901 | 911 | 841.263 | 34.432 |
| 105 | 832 | 845 | 856 | 870 | 880 | 890 | 899 | 909 | 920 | 932 | 950 | 965 | 975 | 901.423 | 36.664 |
| 110 | 891 | 904 | 915 | 929 | 940 | 950 | 960 | 970 | 980 | 993 | 1012 | 1028 | 1039 | 962.037 | 38.322 |
| 115 | 948 | 961 | 974 | 990 | 1002 | 1012 | 1022 | 1032 | 1044 | 1057 | 1078 | 1094 | 1105 | 1024.203 | 40.416 |
| 120 | 1007 | 1022 | 1035 | 1051 | 1064 | 1074 | 1085 | 1095 | 1107 | 1122 | 1143 | 1160 | 1172 | 1086.994 | 42.447 |
| 125 | 1067 | 1083 | 1097 | 1114 | 1126 | 1137 | 1148 | 1159 | 1171 | 1185 | 1206 | 1223 | 1237 | 1149.935 | 43.059 |
| 130 | 1129 | 1145 | 1159 | 1176 | 1189 | 1201 | 1212 | 1225 | 1237 | 1254 | 1276 | 1295 | 1308 | 1215.330 | 46.016 |
| 135 | 1190 | 1206 | 1220 | 1238 | 1253 | 1264 | 1276 | 1288 | 1301 | 1317 | 1340 | 1361 | 1374 | 1278.579 | 47.465 |
| 140 | 1250 | 1269 | 1284 | 1304 | 1318 | 1331 | 1343 | 1356 | 1370 | 1387 | 1409 | 1429 | 1443 | 1345.578 | 49.548 |
| 145 | 1316 | 1333 | 1349 | 1369 | 1384 | 1397 | 1410 | 1423 | 1437 | 1454 | 1479 | 1501 | 1516 | 1412.441 | 51.273 |
| 150 | 1380 | 1399 | 1414 | 1435 | 1450 | 1464 | 1477 | 1490 | 1505 | 1523 | 1549 | 1572 | 1588 | 1479.903 | 53.139 |
| 155 | 1443 | 1461 | 1479 | 1500 | 1516 | 1529 | 1543 | 1557 | 1572 | 1591 | 1620 | 1643 | 1658 | 1546.603 | 55.315 |
| 160 | 1507 | 1526 | 1545 | 1567 | 1584 | 1598 | 1611 | 1625 | 1641 | 1662 | 1690 | 1713 | 1731 | 1614.852 | 57.004 |
| 165 | 1573 | 1592 | 1612 | 1635 | 1653 | 1668 | 1681 | 1696 | 1712 | 1733 | 1762 | 1788 | 1806 | 1684.823 | 59.348 |
| 170 | 1637 | 1658 | 1678 | 1701 | 1719 | 1735 | 1751 | 1767 | 1784 | 1806 | 1835 | 1860 | 1877 | 1754.206 | 61.867 |
| 175 | 1706 | 1727 | 1747 | 1772 | 1791 | 1807 | 1823 | 1839 | 1856 | 1879 | 1909 | 1936 | 1955 | 1826.160 | 63.832 |
| 180 | 1772 | 1792 | 1813 | 1839 | 1859 | 1875 | 1892 | 1909 | 1927 | 1948 | 1981 | 2009 | 2028 | 1895.470 | 66.023 |
| 185 | 1842 | 1864 | 1884 | 1910 | 1929 | 1945 | 1962 | 1980 | 1998 | 2020 | 2052 | 2079 | 2097 | 1965.972 | 66.329 |
| 190 | 1909 | 1931 | 1953 | 1981 | 2000 | 2018 | 2035 | 2053 | 2073 | 2096 | 2130 | 2160 | 2180 | 2039.212 | 69.580 |
| 195 | 1978 | 2002 | 2023 | 2051 | 2071 | 2090 | 2107 | 2126 | 2146 | 2169 | 2204 | 2233 | 2253 | 2111.102 | 70.547 |
| 200 | 2048 | 2072 | 2095 | 2122 | 2143 | 2162 | 2180 | 2198 | 2218 | 2243 | 2279 | 2310 | 2332 | 2184.096 | 72.650 |
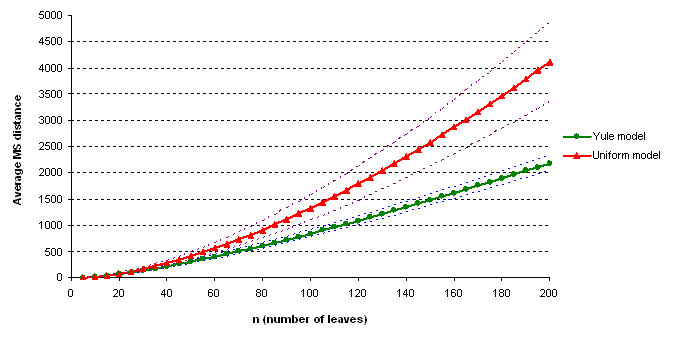
The solid lines show the average values, dashed lines represent double standard deviation intervals (avg�2*sd).