 Visual TreeCmp
Visual TreeCmp
 Visual TreeCmp
Visual TreeCmp
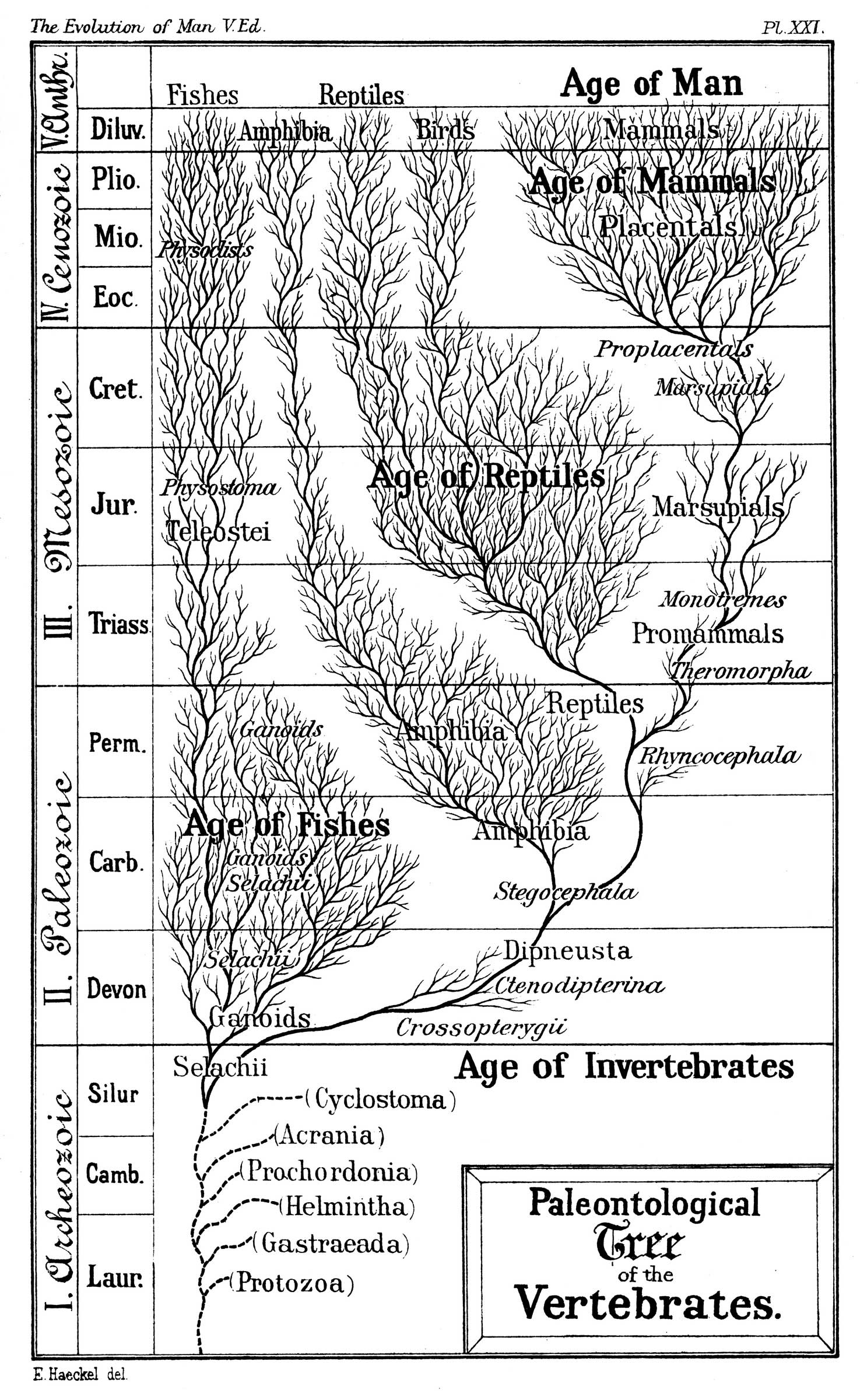
A phylogenetic tree represents historical evolutionary relationship between different species or organisms. There are various methods for reconstructing phylogenetic trees. Applying those techniques usually results in different trees for the same input data.
An important problem is to determine how distant two trees reconstructed in such a way are from each other. Comparing phylogenetic trees is also useful in mining phylogenetic information databases.
The TreeCmp application was designed to compute distances between arbitrary (not necessary binary) phylogenetic trees.
Here is a list of publications related to the TreeCmp and metrics of the authors of the application:
For more details about the MS and MC metrics see related pages: